iNaturalist Introduced Bubble Plot
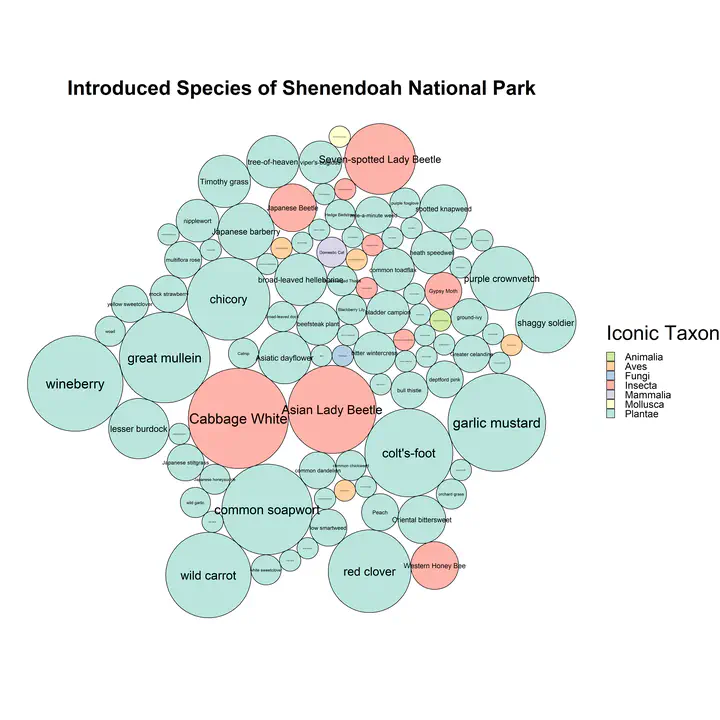
Code to make this figure is below and can be downloaded here: Change the place_id and the place_name to your place of interest. Find your place ID from the iNaturalist search function, when you search your location name in the search bar, like “Fairfax County”, the resulting url has the place name in the url (ex. for the url https://www.inaturalist.org/observations?place_id=738 the place name is 738)
#install.packages("tidyverse", "devtools", "ggraph", "packcircles")
#library(devtools)
#devtools::install_github("hrbrmstr/curlconverter")
library(curlconverter)
library(tidyverse)
library(ggraph)
library(packcircles)
place_id <- 744
place_name <- "PWC"
iconic_name <- NULL
name <- NULL
common <- NULL
api <- paste("curl -X GET --header 'Accept: application/json' 'https://api.inaturalist.org/v1/observations?endemic=false&geo=true&introduced=true&place_id='", place_id, "&quality_grade=research&per_page=200&order=desc&order_by=created_at'", sep = "")
my_ip <- straighten(api) %>%
make_req()
dat <- content(my_ip[[1]](), as="parsed")
for(i in 1:length(dat$results)){
iconic_name <- c(iconic_name, dat$results[[i]]$taxon$iconic_taxon_name)
name <- c(name,dat$results[[i]]$taxon$name)
if(is.null(dat$results[[i]]$taxon$preferred_common_name)){
common <- c(common, NA)}else{
common <- c(common, dat$results[[i]]$taxon$preferred_common_name)}
}
if (dat$total_results > 200){
for (i in 2:floor(dat$total_results/200)){
api <- paste("curl -X GET --header 'Accept: application/json' 'https://api.inaturalist.org/v1/observations?endemic=false&geo=true&introduced=true&place_id='", place_id, "&quality_grade=research&page=", i, "&per_page=200&order=desc&order_by=created_at'", sep = "")
my_ip <- straighten(api) %>%
make_req()
dat <- content(my_ip[[1]](), as="parsed")
for(j in 1:length(dat$results)){
iconic_name <- c(iconic_name, dat$results[[j]]$taxon$iconic_taxon_name)
name <- c(name,dat$results[[j]]$taxon$name)
if(is.null(dat$results[[j]]$taxon$preferred_common_name)){
common <- c(common, NA)}else{
common <- c(common, dat$results[[j]]$taxon$preferred_common_name)}
}
}
}
dat2 <- dat
dat2 <- data.frame(iconic_name,name,common)
dat2 %>%
group_by(common, iconic_name) %>%
summarise(n = n()) -> data
names(data) <- c("group", "iconic", "value")
packing <- circleProgressiveLayout(data$value, sizetype='area')
data <- cbind(data, packing)
dat.gg <- circleLayoutVertices(packing, npoints=50)
dat.gg$group <- rep(data$iconic, each = 51)
# Make the plot
ggplot() +
# Make the bubbles
geom_polygon(data = dat.gg, aes(x, y, group = id, fill=as.factor(group)), colour = "black", alpha = 0.6) +
# Add text in the center of each bubble + control its size
geom_text(data = data, aes(x, y, size=value, label = group)) +
scale_size_continuous(range = c(1,10), guide = F) +
# General theme:
theme_void() +
theme() +
coord_equal() +
ggtitle(paste("Introduced Species of", place_name)) +
scale_fill_brewer(palette="Set3", direction=-1) +
theme(plot.title = element_text(size = 40, face = "bold")) +
labs(fill = "Iconic Taxon") +
theme(legend.text=element_text(size=20), legend.title = element_text(size=40))+
theme(plot.title = element_text(hjust = 0.5))
ggsave(paste("introduced_species_of_", gsub(" ", "_", place_name), ".png", sep = ""), height = 25, width = 25)